Eukaryotic DNA is full of sequences that clearly are not immediately involved with making proteins or RNA. Even as the debate rages on what the meaning is of all this extra DNA, it turns out we can already answer one important question: where did all that stuff come from?
As has been noted, “Life finds a way”. We find organisms living in every ecological niche. And in turn, every organism is itself preyed upon by viruses. It should therefore be no surprise that our genome itself has become a host for things that replicate, mutate and grow, and perhaps “live”.
A lot of our DNA consists of @transposable elements@, @transposons@ or “@jumping genes@”, and these are utterly fascinating in their own right. In addition, they are a wonderful way of illustrating many fundamental things about life and biology. This is because in a very real sense, transposable elements are themselves alive.
So what is this sort of life that hides within us? It turns out it is possible for DNA sequences to be encoded such that they facilitate the further copying of those same sequences. And by this I do not mean the regular replication that happens when cells divide - this is the creation of additional copies on top of that. These ‘transposable elements’ can thus procreate in our genomes.
 Source: Wikipedia user Mariuswalter
Source: Wikipedia user Mariuswalter
Such transposable sequences sometimes succeed in moving around, or inserting a new copy of themselves somewhere else in the genome. And if they manage this feat in germ line cells (@sperm@, @eggs@), this new copy will get passed down to our descendants, where it then might essentially “live forever”.
The circumstances under which this happens are rather rare - our DNA is fiercely defended against such intrusions. But if a sequence manages to copy itself slightly more than once on average, you can see how over timescales of millions of years, a sequence could “go viral”. And it turns out that many transposable elements have - to an outrageous extent even.
As it stands, around 44% of our genome consists of transposable elements or their dead or decaying remnants. In @maize@, this number stands at an astounding 85%.
~~~~~ TRANSPOSON RNA ------------>
/ REVERSE TRANSCRIPTASE
/ ~~~~~~*
=========~~~~~============== DNA =========\/============
TRANSPOSON ENDONUCLEASE NICK
=========~~~~~============== DNA ==========~~~~==============
NEW COPY OF TRANSPOSON
One of the ways in which transposons “copy paste” themselves
Meet the family
There are multiple kinds of transposable elements. Within these kinds there are families, which we can also group in super families. Here we’re going to take a look at two of the most important groups: @L1@ (or LINE1) and @Alu@.
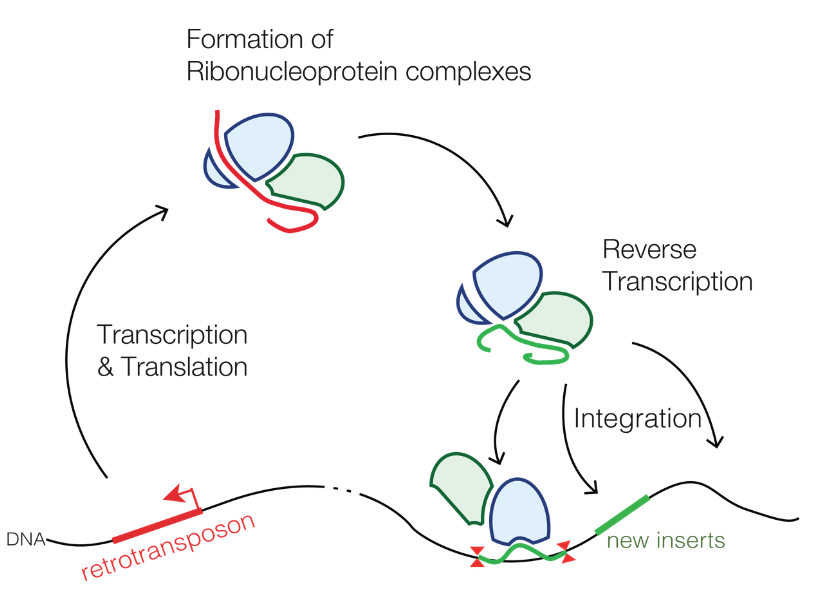
@LINE1@, short for Long Interspersed Nuclear Element 1, on its own comprises around 17% of the human genome. On average, an L1 copy is around 6000 nucleotides long. An active L1 site is able to get itself transcribed to RNA, and this RNA then encodes for two proteins. One of these, an endonuclease, can nick or cut an existing DNA strand. The other protein, a reverse transcriptase, can convert a whole L1 RNA strand into DNA, and insert it in the freshly generated cut. These proteins, with help from existing cell infrastructure, also manage to transport themselves and the RNA to be inserted back into the nucleus, so this is quite an advanced operation
It also often does not work - our chromosomes are littered with truncated and/or corrupted copies of L1. These are unable to replicate themselves further, but they do sit there.
So where did L1 come from? Sadly, it appears we may never know. One of its proteins has no known analogue among all animals. We do know that LINE1 is present in all vertebrates, and we can trace its evolution all the way back to fish, frogs and lizards. By analyzing the difference between L1 among and within these different animals, we know LINE1 has been with us for at least 170 million years.
Over that time, our cells have developed lots of mechanisms to silence L1, so what we observe today is the end of a very protracted war, where L1 is still alive, but barely so. That does not mean it is harmless however.
Having one parasitic layer of life apparently was not enough for nature. There is another main transposon group called Alu. Over 1 million copies (!) of Alu reside in our genome, each around 300 nucleotides long, together accounting for around 10% of our DNA.
Unlike L1, we do however know where it came from - in its 300 nucleotides, we can clearly recognize parts of our very own 7SL RNA gene. This gene is an ancient construct that binds to proteins and helps guide them out of the cell. L1 is shared among all vertebrae, Alu only lives in primates and our closest evolutionary family members, rodents.
Genes carry within them @promoters@ that make the cell’s machinery turn its DNA into RNA, and Alu is no different here. But this is where it gets interesting. Alu is well able to get itself translated into RNA. But to insert a copy of itself, it uses the machinery of L1. Alu does not just “hitch a ride on L1”, it makes creative use of L1 facilities to achieve its goals. In this way, Alu is a parasite on L1 which is a parasite on us.
Transposons may be life form of their own, but if they are, they are living life in extreme slow motion. For example, it is estimated that currently one new copy of Alu gets created every 100-200 births. This does offer some wonderful ways of studying evolution though.
If we look to the other @primates@, we find that we share almost all of the 1 million or so Alu sequences with them. This indicates that the majority of Alu copies happened around or just after the advent of primates. But we don’t share all of them - around 7000 inserts are unique to homo sapiens. These unique inserts offer us a great way to validate the phylogenetic tree of primates. Over time new inserts have happened, but in random places. The number of shared and unique inserts make it possible to trivially determine the relation between species. If one species lacks Alu inserts compared to another one, we know the first is no descendant from the second one, for example.
____1____
/ \
_2__1____ ____1___3
/ \
_2__1_4__ ____1_5_3
/ \
_2__1_4_7 _2_8_1_4__
A species family tree showing accumulation of different transposon inserts over millions of years
In addition, over time, Alu elements mutate. The amount of divergence between inserts also gives us an indication how long ago two species split up.
Intriguingly enough, these mutations also get replicated, until the Alu element is too corrupted to still function. But this copying of mutations allows us to determine the Alu family tree. @AluJ@ is the oldest family, dated to more than 65 million years ago. It is also solidly dead - its copies can no longer replicate, but they still sit there. At 30 million years old, the @AluS@ lineage still has some living elements. The youngest @AluY@ family meanwhile has the highest percentage of active copies.
What do the inserts do?
If a transposable element sets down somewhere in the genome, all kinds of things could happen. Worst case, the transposon decides to liven up the coding sequence of an important gene. Most genes don’t act kindly to lots of new nucleotides popping up. Some genes are absolutely vital, other genes appear to not individually be necessary - if they fail, alternative genes or pathways exist to achieve what that gene did. But often, harm is done.
XXX: interesting article on where TE’s land https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7337890/
In addition, since transposon inserts very much look alike, they can cause problems with chromosome pairing during replication or @meiosis@ (for reproduction). The wrong bits of DNA could stick to each other.
Transposon inserts are associated with many diseases and @cancer@. It is for this reason that our cells valiantly attempt to silence or block transposons.
But it is not all bad news. Many transposon inserts ended up having a function, to the point that removing such inserts may result in a nonviable organism.
It appears that at times, it may be useful to shake up DNA a bit to see what happens. This could be a source of innovation in evolution.
Our immune system also needs very rapid mutation and innovation to quickly evolve antibodies that stick to new and unknown threats. In order to do so, in the DNA of antibody generating cells, variable (V), diversity (D) and joining (J) segments are shuffled in a process called V(D)J recombination. Such cutting and pasting of DNA does not come natural to organisms, but it is the bread and butter of transposable elements. Jawed vertebrae use proteins called Rag1 and Rag2 for this process. @Rag1@ and Rag2 are ancient, and turn out to hail from a transposon family called @Transib@ that to this day survives in starfish, oysters and sea urchins. It appears our immune systems were improved by transposable elements.
So, are these transposons a pest? They surely have been useful in the past. Our genomes are now graveyards of mostly dead transposable elements, and our cells have gotten used to their presence to the point that many of them are now part of the furniture, and we can’t do without them anymore. Mobile genetic elements have also delivered useful innovations, like V(D)J in our immune systems.
My personal theory is that having a genome that so extensively exists of repeats and perhaps non-essential elements is also a form of protection. Any virus invading our DNA is now likely to find itself in a non-coding region, and not impact our lives too much. In a way, all these transposable elements have created a lot of decoy material.
But we also can’t ignore that the still living transposons do cause harm. Having a transposon move is a pretty rare event, but we have a lot of cells. During our lives, transposable elements provably do settle on active genes and cause problems.
These days, we could imagine ridding our genomes of active mobile elements. This would vastly reduce the risk of such harm. But in an abstract sense, this might slow down evolution. But perhaps that is a cost worth paying?
Further reading
- LINE-1 protein localization and functional dynamics during the cell cycle | eLife
- Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates, Genome Res, 2006 Jan
- Transposable elements employ distinct integration strategies with respect to transcriptional landscapes in eukaryotic genomes, Nucleic Acids Res, 2020 May 22
- Frontiers | Evolution of Immune Systems From Viruses and Transposable Elements | Microbiology
- Evolution of the RAG1-RAG2 locus: both proteins came from the same transposon, Biol Direct, 2015 Apr 28
- Alu elements: know the SINEs | Genome Biology | Full Text
- Comparative analysis of Alu repeats in primate genomes, Genome Res, 2009 May
- Structural Variation of Alu Element and Human Disease, Genomics Inform, 2016 Sep 30