The CureVac Vaccine, and a brief tour through some of the wonders of nature
In previous posts I described how the two currently approved mRNA vaccines both use ‘modified nucleosides’ to enhance their efficiency.
Meanwhile, a company called CureVac has created an mRNA vaccine that uses regular, unmodified, RNA. It is currently being tested in large scale trials. In this post I explain what the challenges are with this plain RNA approach, and we reverse engineer quite closely what CureVac has done with regular RNA to (hopefully) deliver an effective vaccine.
I would like to thank Wilson Poon and Martijn Luijsterburg for their extensive help in writing this post. I’d list them as co-authors, but I don’t want to saddle them with the mistakes I’ve surely made. But know that without their help, this page would not be here! In addition, CureVac provided some insights, but they are highly constrained in what they can share because of legal and trial reasons. I do appreciate their help though.
As background, it may be useful to read my earlier article on how mRNA vaccines work, but I’ll also briefly recap things here.
How the BioNTech & Moderna mRNA vaccines work
In short, the goal is to get our cells to produce a single part of the actual SARS-CoV-2 virus. This then activates our immune system, which takes note of what some of our cells are suddenly doing, and of the foreign protein they are producing.
Our immune system learns from these things and develops resistance. When the actual SARS-CoV-2 virus later shows up for real, antibodies and T cells are present to deal with it.
The two approved mRNA vaccines (BioNTech/Pfizer/Fosun and Moderna) and CureVac use small fatty droplets (’lipid nanoparticles’ or LNPs) to sneak mRNA into some of our cells, which then get to work producing the SARS-CoV-2 S protein. Derek Lowe recently wrote about the process of getting the mRNA into our cells and as always with his pieces, it is very much worth your time.
Our cells meanwhile have their own immune mechanisms going on and try hard not to be influenced by foreign mRNA. This could put a rather large damper on the efficiency of mRNA vaccines.
The BioNTech and Moderna vaccines use a chemically modified form of mRNA that is 1) sufficiently different from normal mRNA that it does not set off the cellular immune system (that much) and 2) still looks enough like normal mRNA that it puts our cells to work to produce the S protein.

The CureVac difference
CureVac is a somewhat mysterious place that went through quite some drama in 2020. Donald Trump was rumored to have attempted to buy the company, but this was vehemently denied. There were some weird CEO changes, and at one point the story was that the founder had actually disappeared. Since then, things have settled down a lot.
Unlike BioNTech and Moderna, the CureVac SARS-CoV-2 vaccine candidate CVnCoV (aka ‘zorecimeran’) uses unmodified mRNA, claiming such modifications are not necessary, or even helpful.
I wondered a bit about this and happened to chance on CureVac’s founder Ingmar Hoerr on Linkedin, so I asked him. He responded in public (which is very nice of him):
“For what reason you should have chemical modifications? This makes sense for gene expression for proteins avoiding immune responses as published by Kariko and Weismann. In terms of vaccines I am not convinced to have these modifications”
Initially I did not understand how this would work. I wondered if they had foregone chemical modifications to evade patents, but in a tweet they told me this was not the case.
It turns out they employ some rather impressive alternative techniques instead. They call this “RNActive®”.
Sadly, we don’t yet know how well this approach will work for their SARS-CoV-2 vaccine, but at least in non-human primates, it appears to deliver to a certain extent.
The first readout of the CVnCoV “HERALD” trial is expected somewhere in March or in April.
June 16th UPDATE: The results are in, and they are disappointing.
Also good to note, the CureVac vaccine has been designed to be stable at regular refrigerator temperatures, or even room temperature for a day.
Non-chemical modifications
The mRNA vaccines attempt to get mRNA into our cells to convince them to make copious amounts of the SARS-CoV-2 S protein.
How well this works is clearly a function of at least:
- How much of the mRNA we get into the right part of our cells
- How well the mRNA is picked up by the ribosome (’the protein printer’)
- How long the mRNA survives
- How much attention our immune system is paying
The chemically modified nucleosides from BioNTech and Moderna probably ace the first part - the ‘Toll-like receptors’ pay no attention to modified RNA. This likely makes it easier for the mRNA to survive into the ‘cytosol’, the part of the cell where the magic happens. Note that a lot of the process of how LNPs get mRNA into the cell is poorly understood.
It appears however that CureVac may be compensating for their unmodified mRNA with the three other aspects - a cleverly formulated mRNA sequence may be more easily translated into protein form, and it might also be stable (active) for a longer time.
Finally, natural mRNA does indeed frighten our immune system, which may result in superior development of antibodies and T cells.
According to CureVac, unpurified unmodified mRNA can indeed activate the cell’s immune system too much - they are betting that the CureVac HPLC-purified mRNA hits the sweet spot where their mRNA functions as an “adjuvant” that makes the immune system pay attention.. while not prohibiting the vaccine from reaching the right parts of the cell.
What is in the CureVac vaccine?
At the beginning, things are pretty standard (as in my earlier post). There is a cap, followed by 5’-UTR (untranslated region). As noted earlier, UTRs influence how often, how well and when mRNA gets translated into a protein.

Interestingly enough, the CureVac 5’-UTR has no hits in the big DNA databases. At all. In contrast, the BioNTech 5’-UTR is found easily enough (deriving from ‘Homo sapiens hemoglobin subunit alpha 2 (HBA2)’), but the CureVac one is anonymous. The WHO INN document that describes the CureVac vaccine acknowledges as much and calls this sequence ‘artificial nucleotides’.
A lot of what drug developers do in molecular biology is closer to casting spells than to engineering. Other vaccines appear to have picked a 5’-UTR from nature that is known to work well. Such sequences are often hundreds of nucleotides long. CureVac has instead inserted a very brief sequence that appears to consist of the bare minimum to make things work.
Wilson Poon and Martijn
Luijsterburg helped me to fully reverse
engineer the very brief 5’-UTR sequence: GGAGA AAGCUU ACC. Many thanks!
Starting at the end, ‘ACC’ is a near-mandatory part of the Kozak sequence, a sequence that tells the ribosome that the start codon is next, and that the protein coding region is about to begin.
AAGCUU before that is mentioned in CureVac
documents as a
flanking cloning
site.
This is probably an artifact of a ‘subcloning
technique’. So it likely is
not there because it is biologically helpful, but because it has to be to
there to enable the bacterial HindIII restriction enzyme
to cleave the DNA in the right place during the production of the mRNA
vaccine.
This leaves us with start of the 5’-UTR, GGAGA. The production process of
mRNA vaccines starts with DNA that then needs to be converted to RNA. This
conversion is performed by a protein we borrowed from the T7 phage, a
bacterial virus.
T7 polymerase needs a leader sequence before it starts its noble conversion
work for us, and this sequence
is TAATACGACTCACTATAG|GGAGA, where the
first part is the leader and the second part is the first DNA to actually be
transcribed to RNA. And lo, GGAGA is what we see at the start of the
vaccine.
Wrapping it up, CureVac 5’-UTR is very minimal and mostly appears to contain “technical” artifacts from restriction and replication enzymes, plus the mandatory Kozak sequence.
A small aside on ‘restriction enzymes’
In various places of the CureVac vaccine we find evidence of the use of restriction enzymes. These things are fascinating, and we’ve borrowed them from the bacterial realm to cut DNA for us.
A restriction enzyme recognizes a specific DNA sequence, and then cuts it. Some of these enzymes cut in a very specific place close to or within the matching sequence, and that makes these enzymes very useful for genetic manipulation activities.
Bacteria sport a lot of these restriction enzymes. It turns out that we aren’t the only ones getting infected with viruses, bacteria also suffer from this problem. Bacteria have been around for four billion years now, and it appears they’ve been doing battle with viruses for a lot of that time.
Whenever a bacterial virus (also known as a phage) injects its DNA in a bacterium, restriction enzymes stand ready to cut this DNA into pieces. By “matching” on many short DNA sequences, they’ll almost always match somewhere in the viral genome, where they can then do damage.
This is of course nice, but how do bacteria prevent their restriction enzymes from cutting up their own DNA? It turns out they have a second set of enzymes going round that ‘paint’ over the areas that would be attacked by the restriction enzymes, thus protecting them. A freshly arrived phage chromosome does not yet have such paint (’methylation’) to protect it.
The key of course is not to paint a bacterial virus chromosome before the restriction enzymes can cut it up, and this sometimes fails. Oddly enough, once such a phage has been painted like that & takes up residence in a bacterium, when it then multiplies, its offspring will receive protective paint as well and be able to infect yet more (identical) bacteria!
Many restriction enzymes are unreasonably precise and there is a strong suspicion that more is going on than meets the eye. But no matter the reason, restriction enzymes are trusty tools of the trade in the world of molecular biology.
Back to the vaccine, where we encounter the S protein itself
There are 64 codons (4*4*4), but only 20 amino acids. Many amino acids therefor have 4 or even six ways of “writing” them in RNA. This leaves a lot of room for changing the RNA sequence, without changing the actual protein.
All vaccines have done so in a grand way, at the very least because it is known that stuffing as many C and G nucleotides in there as possible is a way to enhance mRNA stability and improve protein expression.
The original SARS-CoV-2 RNA has a GC content of 37%. CureVac raises this to 63% while BioNTech clocks in at 56%.
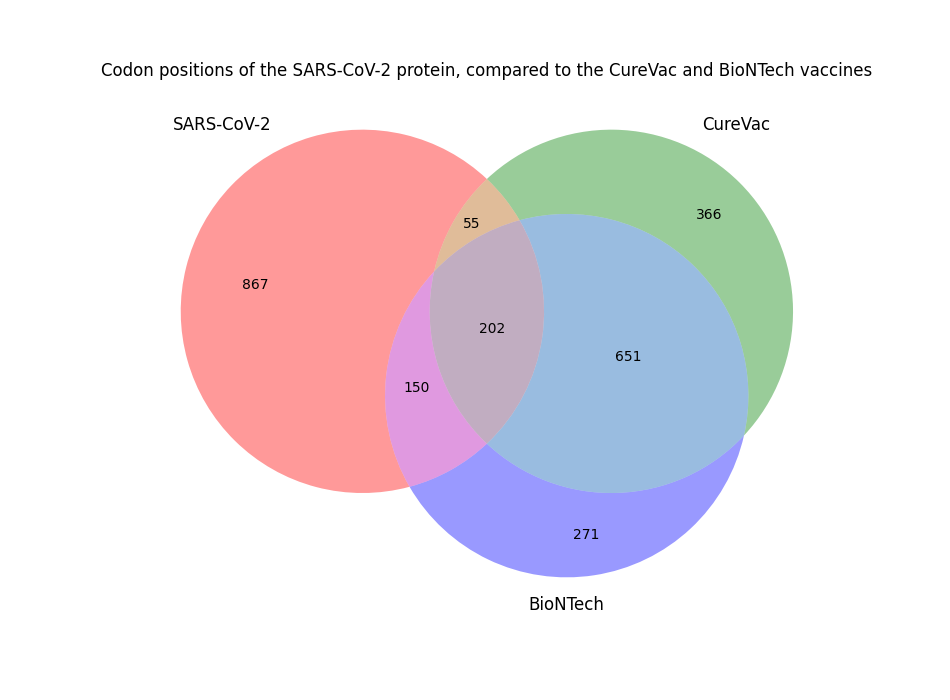
There are 1274 codon positions in both vaccines and in the original SARS-CoV-2 S protein. In this Venn diagram, we can see that 202 of these are the same in both vaccines and the original virus.
We can also see that 366 changes are unique for CureVac, whereas BioNTech has 271 unique modifications. Finally, 651 modifications are common to both vaccines.
Overall it looks like the CureVac version is modified more heavily.
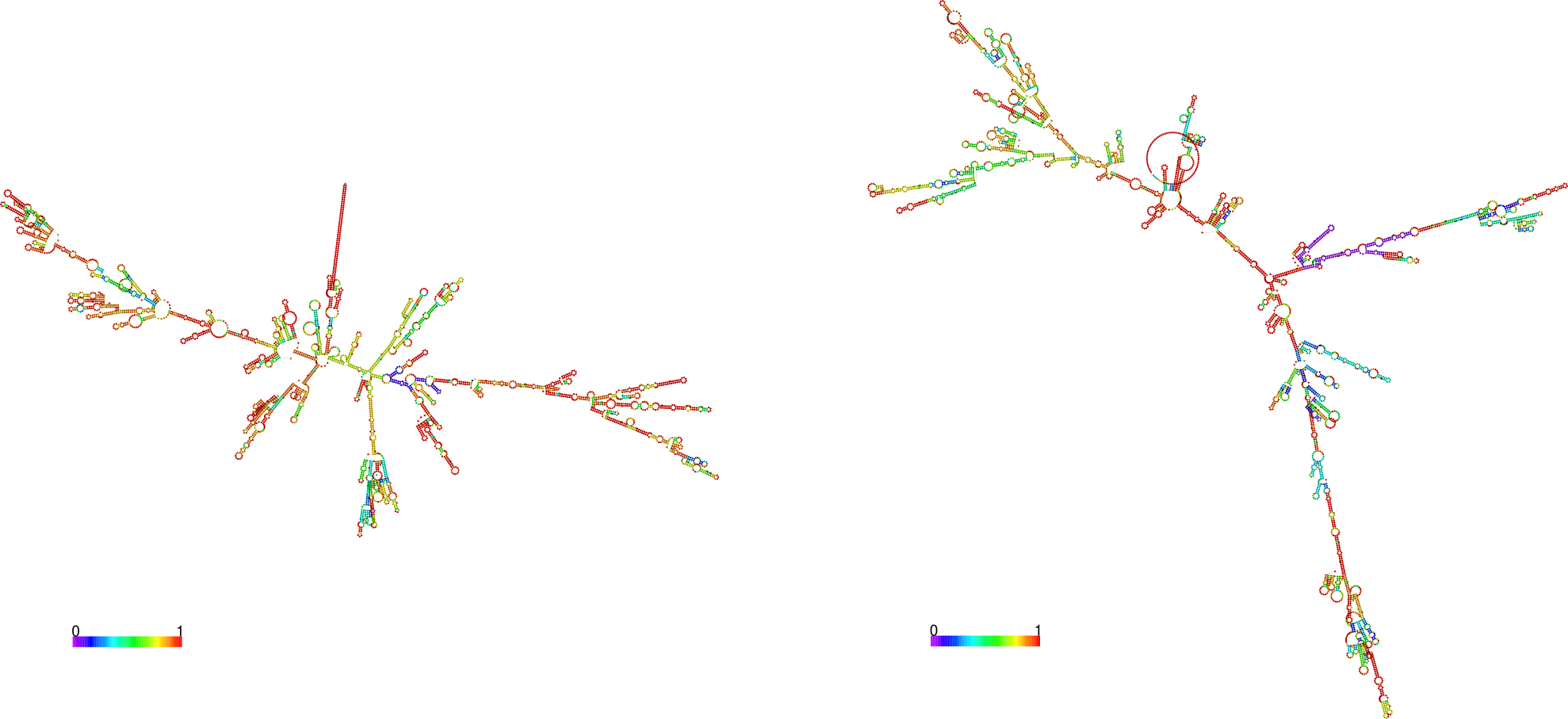
RNA clings to itself depending on its sequence. It is possible to calculate the likely ‘folded’ shape of a string of RNA. When we ask the impressive RNAFold server of the University of Vienna to fold the two vaccines, we end up with the pictures above.
The CureVac sequence on the left appears to fold a lot “tighter” than BioNTech on the right. I’ve asked around if this could be significant, and I am told it might or it might not be. But at least we have a pretty picture to look at.
The CureVac vaccine contains the “2PP” modification that is also present in the Moderna and BioNTech vaccines, but not the further modifications we find in the Janssen and Novavax vaccines.
The 3’-UTR
Not only is there an Untranslated Region in front of the actual protein description, there is one at the end as well, and this is then called the 3’-UTR.
The CureVac 3’-UTR is described as ‘3839-3912: untranslated region (3’ UTR) comprising the following element: Human alpha-globin 3’ UTR sequence element (3861-3904)’.
What this means is that the 3’-UTR is 74 nucleotides long, of which 44 nucleotides in the middle were sourced from a piece of human DNA.
The first 23 nucleotides GGACU AGUUA UAAGA CUGAC UAG are of unspecified
origin, but if we use the venerable ‘BLASTn’ search, we find many partial
hits within mRNA sequences found in other organisms, mostly in the
Odontomachus brunneus ant species. This may well be a coincidence however.
The ACU AGU part in there hails from SpeI/AhII/BcuI restriction enzymes,
as mentioned above.
The middle ‘human’ bit of DNA is recognized by BLASTn as hailing from the 3’-UTR of Homo sapiens alpha-2 globin (HBA2) gene.
Meanwhile, the last nucleotides GAGAU UAAU show no hit on BLASTn, and I
have no idea what this might be.
Poly(A)
Human mRNA typically ends in an AAAAA-sequence. This sequence protects the mRNA from degradation. Over time, trailing A’s are lost, and once the Poly(A)-tail is gone, the mRNA is quickly eliminated.
This feature is present in both vaccines we have the sequence for.
Poly(C)
Now this is where it gets interesting. As noted, the name of the game for mRNA vaccine efficiency is to get the cells to produce sufficient SARS-CoV-2 proteins. To do so, enough mRNA must make it into the cytosol, and it should also be picked up often enough by the ribosome. And then it has to stay around long enough.
Given that unmodified nucleosides are probably less likely to evade our immune system, the CureVac vaccine must compensate elsewhere.
Here we encounter the first region where CureVac has done something different, a Poly(C) stretch (aka a bunch of C’s).
Research into C-rich motifs within RNA is still rather fresh, but signs are that when αCP (or poly(C)-binding protein (PCBP)) binds to such a region, [it can enhance the stability of the mRNA](s poly(C)-binding protein (PCBP)), as well as further the expression of it.
Histone stem-loop
Human chromosomes are huge. While a bacterial chromosome may consist of a few million base pairs, the human genome clocks in at over 3 billion of them. And because chromosomes are present in duplicate, this means that a nucleus hosts over 6 billion nucleotides.
A human nucleus is around 10 μm in diameter and contains around 2 meters of DNA in total. Getting two meters (6 feet) of DNA into such a tiny volume requires some work. If you’ve ever tried to store cables in a box, you’ll know how well that goes.
Human (and animal and plant) DNA is stored with the aid of “histones”. The complexity is huge, but the summary of the summary is that histones band together to form spindles around which DNA gets looped. This storage combines stability with (selective) availability.
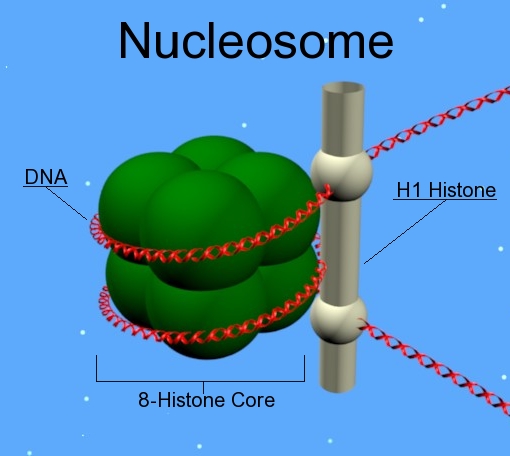
Source: Wikipedia
Copying a human cell takes a lot of time, and specifically the “S phase” in which the new DNA gets created and stored lasts for many hours.
Crucially, during this S phase, the DNA is busy being copied, and not a lot of new RNA is generated while that happens. However, during the replication, a steady supply of a huge number of histones is required, so new DNA can be stored correctly.
In other words, what is needed are highly stable mRNA sequences that will continue to generate histones for hours and hours.
This sounds just like what we need for our vaccines!
The mRNA sequences that generate histones do not have a poly(A) tail, but they do have a ‘histone stem-loop sequence’ at the end. This likely ensures their longevity.
In what proved to be a successful experiment, CureVac discovered that you can add both a poly(A) tail AND a histone stem-loop to mRNA, and that these mechanisms have an additive effect on mRNA stability.
“The present invention is based on the surprising finding of the present inventors, that the combination of a poly(A) sequence or polyadenylation signal and at least one histone stem-loop, even though both representing alternative mechanisms in nature, acts synergistically as this combination increases the protein expression manifold above the level observed with either of the individual elements. The synergistic effect of the combination of poly(A) and at least one histone stem-loop is seen irrespective of the order of poly(A) and histone stem-loop and irrespective of the length of the poly(A) sequence.” - from this CureVac patent.
Summarising
The CureVac mRNA is not chemically modified. This may mean it has a harder time reaching the right part of the cell, since the immune system does not like foreign mRNA.
Inside the CureVac mRNA sequence we find various mechanisms that likely increase both the activity and longevity of the vaccine mRNA. Relatively speaking, the CureVac sequence is more modified than that found in the other vaccines, and it contains two additional sequences that might make a big difference: the poly(C) sequence and the histone stem-loop.
Additionally, the unmodified nature of the mRNA could also be a blessing: by alerting our immune system, a more vigorous response may be elicited.
Soon we will learn the outcome of the phase 2/3 “HERALD” trial, which will hopefully tell us if the non-chemically modified CVnCoV mRNA vaccine is efficacious against SARS-CoV-2.
June 16th UPDATE: The results are in, and they are disappointing.
Final note
If you like this kind of “semi-popular” science writing, you can hire me!.
