On the Pan-Genome
In this post I want to shine a light on an important development within the world of DNA which shows how much DNA and the world of computing are now intertwined. It turns out we share important problems, like in this case: file formats.
Note: I expect to be posting more DNA material as part of my efforts to write a book on this fascinating subject. If you enjoy reading about DNA, you might like the links/presentations/videos found on this page. Feedback is welcome on bert@hubertnet.nl or @bert_hu_bert!
It is well known that the “human genome” is now fully sequenced (‘read’), and that we can download all of it easily enough. It turns out neither of these statements is true!
When we say “the human genome” is sequenced, this is actually DNA from a mix of people, so the “one true sequence” online represents no single individual. In addition, by no means does this genome capture the variability of actual people out there.
Because the dark secret is.. although everyone is different, the chosen DNA file format ("FASTA") has no way to encode that. It encodes a single string of letters as the one true genome. This means that if variants are present only in some people, most of these will not make it to the official human genome collection (some specific variations are supplied as separate files though).
Here is a FASTA sample from the human genome, specifically from the mitochondrial DNA:
>MT dna:chromosome chromosome:GRCh38:MT:1:16569:1 REF
GATCACAGGTCTATCACCCTATTAACCACTCACGGGAGCTCTCCATGCATTTGGTATTTT
CGTCTGGGGGGTATGCACGCGATAGCATTGCGAGACGCTGGAGCCGGAGCACCCTATGTC
GCAGTATCTGTCTTTGATTCCTGCCTCATCCTATTATTTATCGCACCTACGTTCAATATT
ACAGGCGAACATACTTACTAAAGTGTGTTAATTAATTAATGCTTGTAGGACATAATAATA
ACAATTGAATGTCTGCACAGCCACTTTCCACACAGACATCATAACAAAAAATTTCCACCA
AACCCCCCCTCCCCCGCTTCTGGCCACAGCACTTAAACACATCTCTGCCAAACCCCAAAA
ACAAAGAACCCTAACACCAGCCTAACCAGATTTCAAATTTTATCTTTTGGCGGTATGCAC
TTTTAACAGTCACCCCCCAACTAACACATTATTTTCCCCTCCCACTCCCATACTACTAAT
CTCATCAATACAACCCCCGCCCATCCTACCCAGCACACACACACCGCTGCTAACCCCATA
So how does it work? FASTA is literally a text file with DNA letters in there, mostly one file per chromosome. To find a gene, we note its offset in the file, and then refer to its ’locus’ as ‘1243553 on chromosome 1 of GRCh38’. Periodically new releases of the genome are made available, and then all numbers change. For this reason, large fractions of researchers are still stuck on GRCh37, because that is where all their numbers point.
The pan-genome
There is significant variation between humans, with even the size of our DNA varying by over 1%. For various bacteria, attempts have been made to capture not just the DNA of an individual population but also that of different populations (or strains). This then captures not just a single genome but also genes present in some, but not all, individual organisms. This broader assembly is called the ‘pan-genome’.
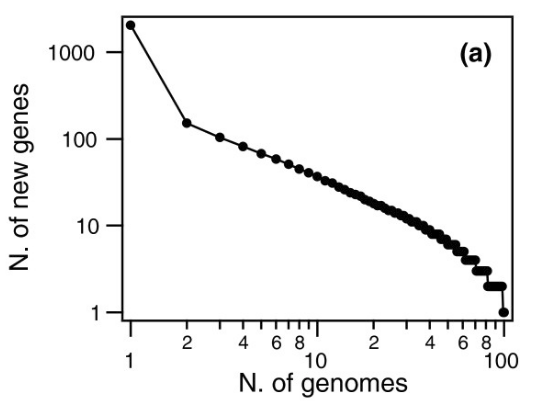
For example, when the bacterium S. pneumoniae was studied, the following was found:
This graph means that when the second S. pneumoniae strain was sequenced, more than 100 additional genes were found. The third one added around 100 more, and so on, until after the 50th organism when almost no more new genes showed up.
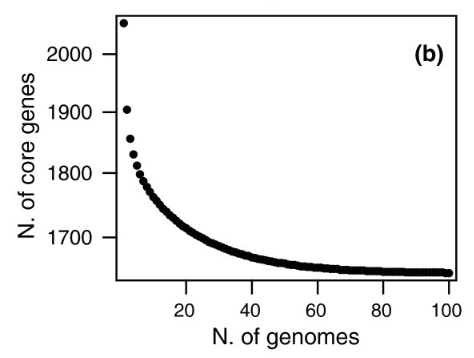
Meanwhile, the inverse also happened, the group of ‘core’ genes, present in all S. pneumoniae copies, went down:
As you can imagine, there is no way to capture all this variation into one linear DNA file.
Graphs
Instead of maintaining the fiction that there is “one true DNA”, a better way would be to store chromosomes as graphs. Such a graph could then encode elements that are common to all individuals as a straight line, but then split up in places where multiple variants exist:
TTCGTTAT
/ \
ACCGTCCTGATTCAAT __TTGCAGTGTCCAA
\ /
CCTGTA
Or perhaps some parts of the genome are not universally present:
TTCGTTAT
/ \
ACCGTCCTGATTCAAT----------TTGCAGTGTCCAA
This denotes an organism where some individuals have this TTCGTTAT sequence, and others do not.
So why don’t we do this?
Despite valiant efforts by several groups, the world of DNA files remains stubbornly flat. One reason is that biologists/bioinformaticians are very attached to being able to address a piece of DNA as a number, the previously mentioned ’locus’. This is embedded deeply in all kinds of processing pipelines, to the point that even adapting to a newer genome release (with different numbering) is considered very painful.
Any kind of ‘graph’ storage makes it harder to point to a specific DNA location with a descriptor, let alone a single number. In the trivial examples above, it may be possible to create an address that encodes that a mutation lives on the ’top’ branch of the graph, based perhaps on blessing one DNA path as the ’true’ one.
It turns out however that real “pan-genomes” not only have alternate pieces of DNA, the alternate pieces also have optional elements, which again have alternate sequences on top of them etc. In addition, some bits of DNA are the same in different individuals, except they are inversed, for which it makes sense to have a dedicated notation.
So a generic DNA graph file format should support arbitrary linkages and nestings of DNA variants, with named branches, which makes addressing where a locus lives a non-numerical affair.
What is happening?
It is clear that pretending there is ‘one DNA’ is no longer tenable. Third generation sequencing techniques can now read very long stretches of DNA in one piece, and such pieces very clearly show structural variations in chromosomes not captured by linear files.
For five years, people like Erik Garisson and the vg team have been slaving away at creating a whole graph-aware software stack, one that does all the heavy lifting to process all kinds of DNA information into the brave new world of non-linear DNA storage. Their work includes exciting new file formats that can encode DNA graphs efficiently.
Graph-aware processing requires far more CPU and memory. And while the rewards can be enormous, life is far more complex in this world. But it is obviously the right thing to do.
Alternatives
Very recently, one of the many geniuses in the world of DNA software, Heng Li released a software stack that also performs graph DNA processing.
In his stack, he uses a new text based format called rGFA, short for “reference Graphical Fragment Assembly”. One of the major advantages of rGFA is that it allows for stable references to locations (“coordinates”), but crucially, only as long as the ‘reference DNA graph’ is stable. If a later addition of more information leads to a change of shape in the DNA graph, the ‘coordinate’ of a location will again change.
In addition, the new format comes with limitations having to do with small scale variations.
In short, Heng Li’s work is likely to be easier to get used to, and his software is also faster, but the end result is not as generic as it could be, and again locks down “official sequences” because this keeps the simple coordinates stable.
What will happen now?
As an outsider, I’m amazed how linear DNA processing still is. I can appreciate how much work it is to change the status quo, especially with so much of medicine still on the previous version of the human genome.
I have also seen however how often convenient solutions, that have shortcomings, win out over better solutions that are initially less convenient.
So it appears that Erik’s worries about the future of graph-based pan-genomes are justified.
We could easily end up on a suboptimal but easier to use solution - which is not far removed from the current linear DNA files!