A Science Experiment: part 1
So, I think I may have discovered something interesting in biology! Professional scientists know this feeling all too well. Exhilarated that it looks like you might be the first person ever to know something, but worried sick that it might not be real. Also, you might be fooling yourself – and you are the easiest person to fool.
And even if it is real, does it even mean something? Or did you effectively discover that hot things are not cold? Another worry is finding out some Russian genius scooped you … in 1983.
This is actually how most of my “discoveries” so far have ended. I count myself rather lucky if the actual discovery happened in the last 100 years. There is already some limited but real joy in this though, as described in this discussion between W. Daniel Hillis and Richard Feynman:
We worked out a model of evolution of finite populations based on the Fokker Planck equations. When I got back to Boston I went to the library and discovered a book by Kimura on the subject, and much to my disappointment, all of our “discoveries” were covered in the first few pages. When I called back and told Richard what I had found, he was elated. “Hey, we got it right!” he said. “Not bad for amateurs.”
Anyhow, the stage of potentially having discovered something is scary. And it is doubly scary if you are an outsider to the field of your discovery. Odds are massive that you are deluding yourself.
Now, traditional academic environments have all kinds of mechanisms to help people find out if what they are doing makes any kind of sense. Seasoned professors and PIs stand ready to tell you your idea is bunk, for example. “Your manuscript is both good and original; but the part that is good is not original, and the part that is original is not good.”
Meanwhile, scientists frequently present what they are working on in colloquia, symposia, lunch meetings and other interactive settings. All this provides valuable feedback, and possibly encouragement.
Sitting here in my home office, I mostly lack this academic peer group. The good news is that there are some great people on Twitter to spar with from time to time – which is already very useful. But I need more.
So, as an experiment, I’m going to document my discovery process here, both to solicit feedback and to create some pressure on myself to actually get this thing done!
So, what am I working on?
If you want to beef up a bit on DNA and biology, and come from a computing background, I have some relevant material on offer:
- Amazing DNA - DNA seen through the eyes of a coder (or, If you are a hammer, everything looks like a nail)
- DNA: The code of life - two hours of slides and video
- What is Life
An intermezzo
Biology is utterly fascinating. Earlier I wrote the following:
Imagine a flashy spaceship lands in your backyard. The door opens and you are invited to investigate everything to see what you can learn. The technology is clearly millions of years beyond what we can make. This is biology. That’s why it fascinates me so much. Life is billions of years old technology, and we can explore it to see how it works!

Take me to your leader
Almost everything that we as humans have ever thought of existed in nature for millions of years already: information storage, complete chemical factories, signal processing, image processing, measurement and control technology, artificial intelligence (but then the real thing). And invariably we discover that the technology of nature is many times more powerful than what we ever invented as humans.
We are surrounded by the end result of 4 billion years of evolution. But we still barely understand how it all works.
In effect, it is like living in a 4 billion year old crime scene, in which we are trying to figure out what happened. Clues abound. There is old life, there is younger life. Some inventions were made many times (vision), others only once (photosynthesis). Some things vary wildly, other things (like reliable DNA copying speed) are astoundingly constant among the entire tree of life – hinting at something fundamental going on.
Some life is incredibly complex (elephants, mushrooms, human beings), other parts are simpler (bacteria). And then there are semi-living things that on the face of it are even simpler (bacterial viruses, phages).
Yet all of life exhibits patterns, patterns which might help us discover profound things about the nature of life.
Forensic traces
Earlier I spoke about life as a crime scene, by which I of course do not mean that life itself is criminal. However, the science of forensics is applicable here – every contact leaves a trace.
In our DNA, we find heaps of flotsam and jetsam we know originated as viruses, ages ago. Some of these bits of DNA have managed to continue to copy themselves, even if at very slow rates. Slow enough that we can tell from the primate family tree exactly how it must have split up - some of these copied DNA snippets only appear in certain lineages, and not in others.
In this way, life has left tangible fingerprints on the otherwise undated DNA record.
DNA & its one-way problem
DNA is digital data storage with analogue retrieval technology. We can store arbitrary data in DNA, and it serves that purpose very well. Every human being contains around 750 megabytes of unique DNA, for example. We routinely recover DNA that is tens of thousands of years old. It is good stuff.
The analogue retrieval is where DNA is a lot different from (say) a hard drive. Molecules with specific shapes can activate bits of DNA very effectively, for example. This could perhaps be likened to Content-addressable memory. This allows life to divvy up the right bits of DNA without any seeking or addressing.
In other ways however DNA is remarkably recognizable. And despite 4 billion years of evolution, it shares a major limitation with our digital technology: it can only be read in a single direction.
If you play an MP3 file, your computer or phone will read the file linearly. Everything is optimized for that. If you try to play back music in reverse, or perhaps want to rewind, you’ll find that your computer really can’t do that. In reality, it will jump back a lot in the file, and from that point on will read the file in the forward direction again.
In a hard drive this is even more recognizable - it only rotates in one direction. You can’t reverse that. If you want to read a file in reverse order, the drive will read a bit of data in the forward direction, jump back a whole bit, and again read something in the forward direction.
In computing, this rarely is a problem, since we store all our files sequentially – they all want to be read from beginning to end.
Nature however decided to be different. DNA consists of the famous double helix. The double aspect means that all of DNA is present twice. So if we want to store ‘ACGTTCA’ in DNA, in effect it will look like this:
<-------
ACGTTCA
|||||||
TGCAAGT
------->
This shows the two strands of DNA, where each A finds itself opposite a T and every C is attached to a G. If damage occurs, it can easily be repaired, because the opposite side forms a template to attract replacement nucleotides. A/T and C/G pairs can each be compared to north/south pole magnets, they attract each other strongly.
But also note the two arrows - DNA has directionality, and both halves are read in opposite directions.
Genes can actually live on both sides. Some very artistic genes even manage to overlap, meaning both sides of the DNA are simultaneously part of two different genes.
Okazaki fragments
Recall that DNA can only be read in one direction. Normally this is fine, since a gene is read in the ‘forward’ direction, regardless of which strand it is on. This can be compared to playing an MP3 file, and not playing it in reverse.
However, when DNA gets copied.. both halves must be copied simultaneously, and in one (spatial) direction. This means that one half should be read in reverse during the copying process. And here’s the rub. Nature can’t do that. Much like you can’t spin a hard drive in the opposite direction, nature can do nearly zilch to DNA in the wrong direction.
So what happens during copying.. is truly amazing.
Source, wehi.tv, Drew Berry
In this video, DNA to be copied streams in from the left, and is then split into two strands by the blue sponge ("Helicase"). One strand then moves downwards, and in streaming fashion, this single strand gets augmented into double-stranded DNA. This is all very straightforward. This is the strand that moves in the natural direction.
But the other strand! You can see that a whole circus goes on. The loop you see extrude & then get pulled in again is 100% comparable to what happens in a computer when we try to read a file in the reverse direction - chunks get read in the forward direction, then copied in reverse, and the results gets concatenated.
In DNA we call these chunks Okazaki fragments.
Without going into too much detail, one can imagine that the simple replication process of one strand might might lead to different errors and mutations than the very complicated process of the other strand. The mistakes in copying are guaranteed to be different.
Over time, these differences might lead to traces in our DNA. This can be compared to how we’ve been able to reconstruct if vehicles in Pompeii drove on the left or the right side of the road by cleverly analysing ruts in the road and scuff marks on corner pavements.
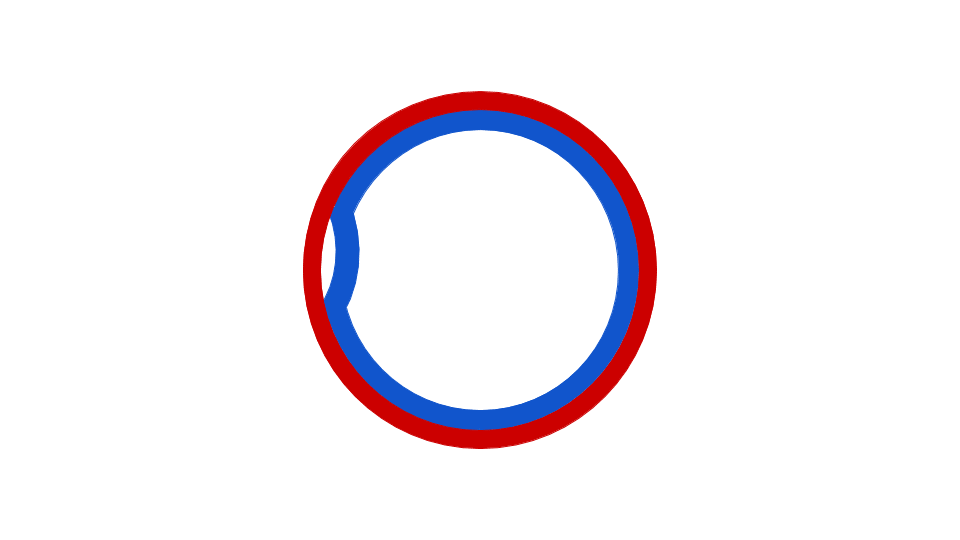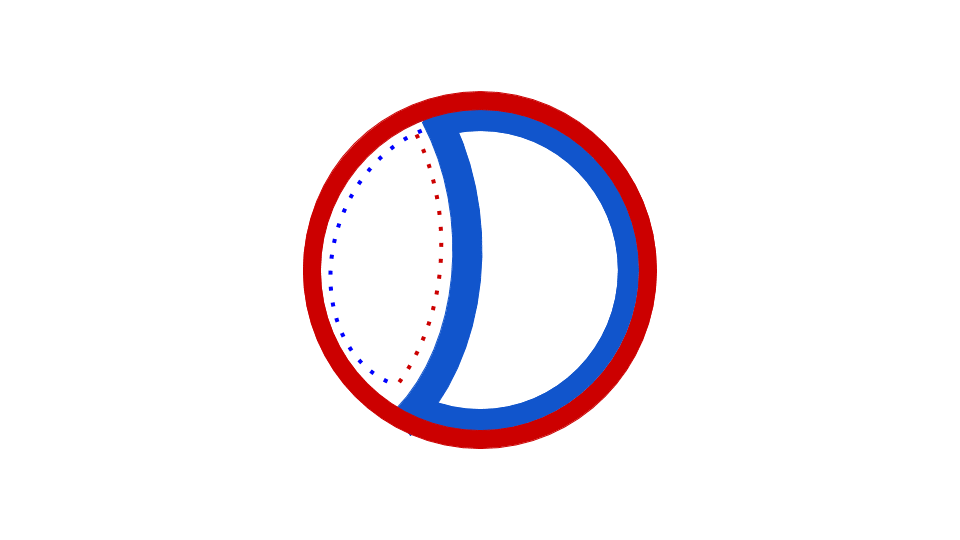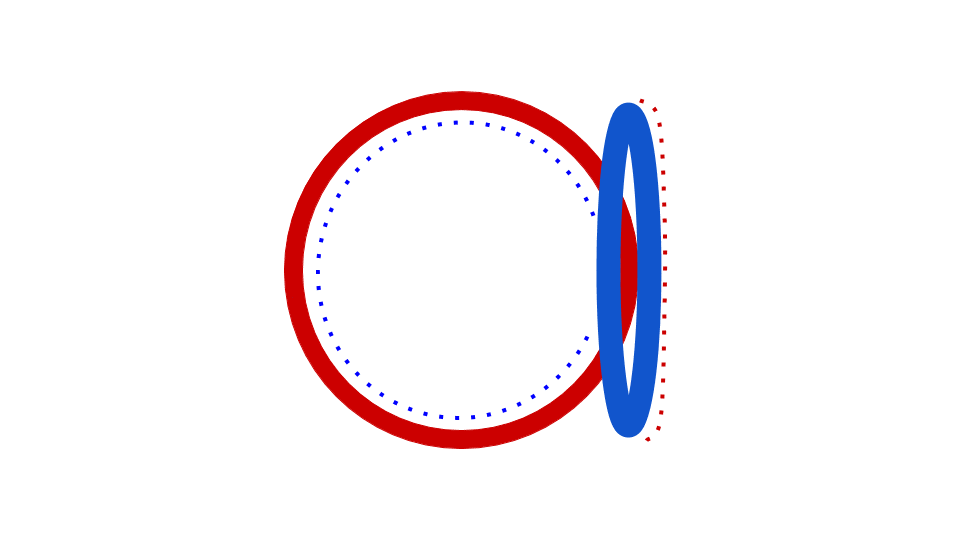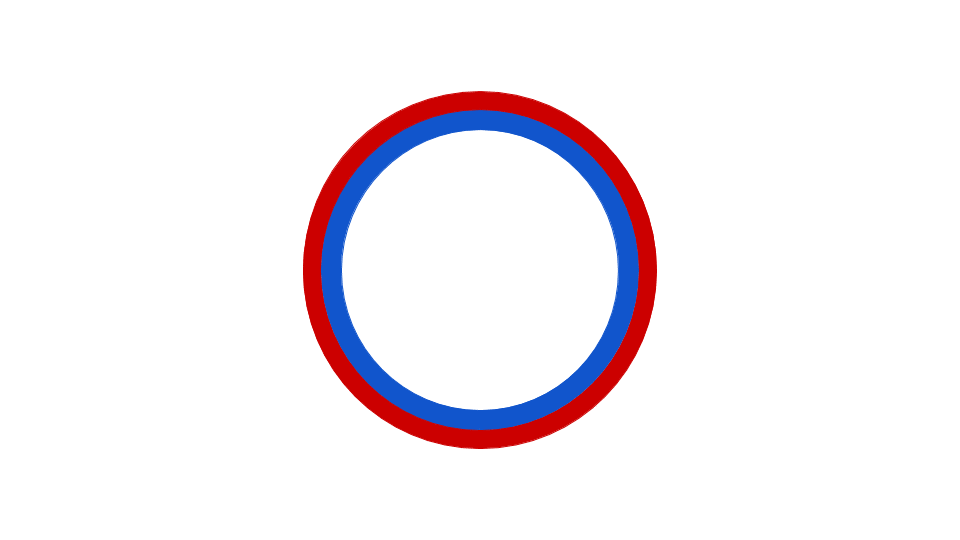
Eubacterial DNA copying commences at the evocatively named Origin of Replication, which has a fixed place in chromosome. Observe that copying happens in two places simultaneously, once in the top half, once in the bottom half of the chromosome. Source: Wikipedia user Catherinea228.
It turns out that in some organisms, notably eubacteria, specific regions of the genome always get copied in the same direction. This means that over time, even tiny differences in the copying process of both strands will add up to sustained effects.
Bacteria can copy themselves every 20 minutes. This means that there is ample chance over the millennia to accrue slight differences between the two replication processes. And lo, this has happened.
DNA consists of four “nucleotides” we call A, C, G and T. Fascinatingly, on each full strand of DNA, there are always equal amounts of G and C. Some biologists tell me this is is so trivial that it doesn’t even need to be studied, other researchers come up with complicated mathematical frameworks to explain why the numbers of C and G nucleotides match up so closely.
It turns out that most bacteria indeed do have very similar amounts of G and C (on each strand), but that one half of the chromosome has an excess of G and the other half has an excess of C. This phenomenon is known as GC skew:
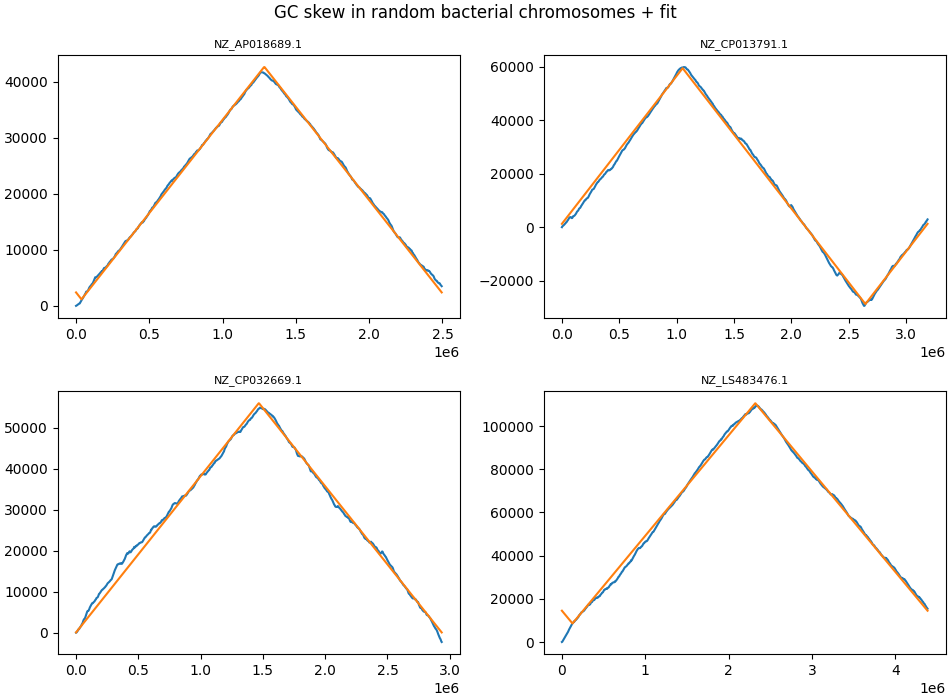
This y-axes of this plot show the cumulative excess of G over C as we go over chromosomes from beginning to end. The bacterial chromosomes plotted here vary in size from 2.5 million nucleotides to 4.5 million nucleotides (this is on the x-axis).
These are some of the straightest lines you will ever see in biology. The orange lines are a simple fit, and these deliver parameters for all of the ~23000 bacterial chromosomes we have data for.
In biology, it is quite rare to get data where the graphs are so striking.
Next up
By analysing all 23000 chromosomes for which we have data, certain patterns become clear that may help us a tiny bit further along the path of understanding why GC skew happens, and why it is different in some classes of bacteria than in others. And this is where I think I may have made a small but hopefully relevant discovery.
In part two of this series I continue the story, and delve into what these 23000 chromosomes are doing, and what patterns emerge.
I’ve added a comment section to this post, please feel free to ask any and all questions you have there! Suggestions, questions, feedback etc is also very welcome via email bert@hubertnet.nl, or to @bert_hu_bert. If there is interest I can also share the software and data set. Please let me know!
